Hamm MO, Wright RC
currently in development
This web app and open source software package for the R programming language was built to facilitate exploration of the naturally occuring sequence diversity for your favorite gene or gene family. We hope this web app will help plant biologists without bioinformatics experience access the 1001 genomes project’s rich catalog of natural variation and formulate hypotheses and identify new alleles in their favorite gene family.
The 1001 genomes dataset was collected with the intention of genome scale analyses, such as genome wide association studies, in which a particular phenotype is quantified for all of the accessions and sequence variants that are associated with phenotypic variation are identified. In my recent work, I have utilized this rich dataset in a different manner to measure the effects of coding sequence variation on protein functional variation and phenotypic variation. This work has expanded the sequence-function-phenotype map for the auxin-signaling F-box gene family, and this web app will facilitate similar analysis other genes, gene families, and genomic regions.
Tabs within the webtool will help you:
- calculate nucleotide diversity for a set of genes,
- plot the nucleotide diversity of missense and synonymous variants,
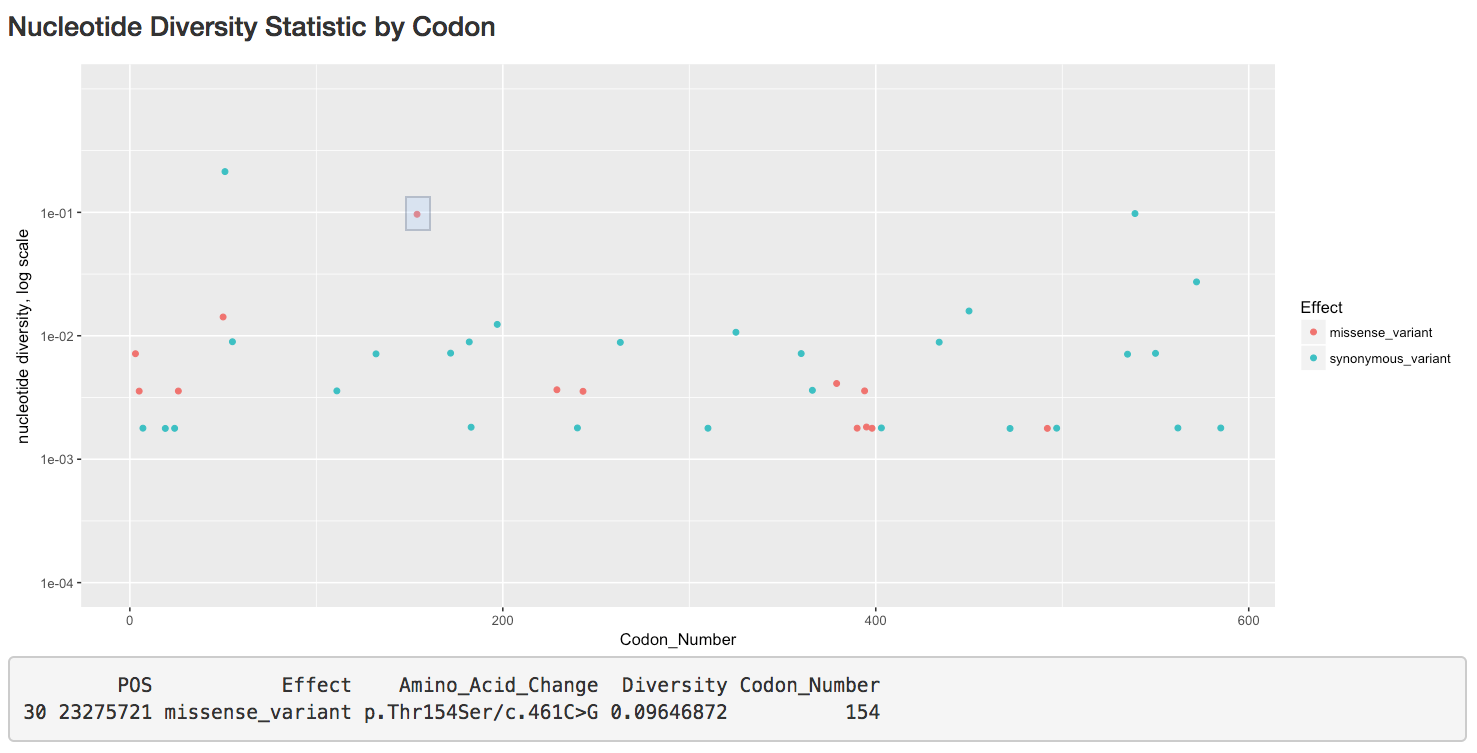
- map accessions containing variants in a gene or set of genes,
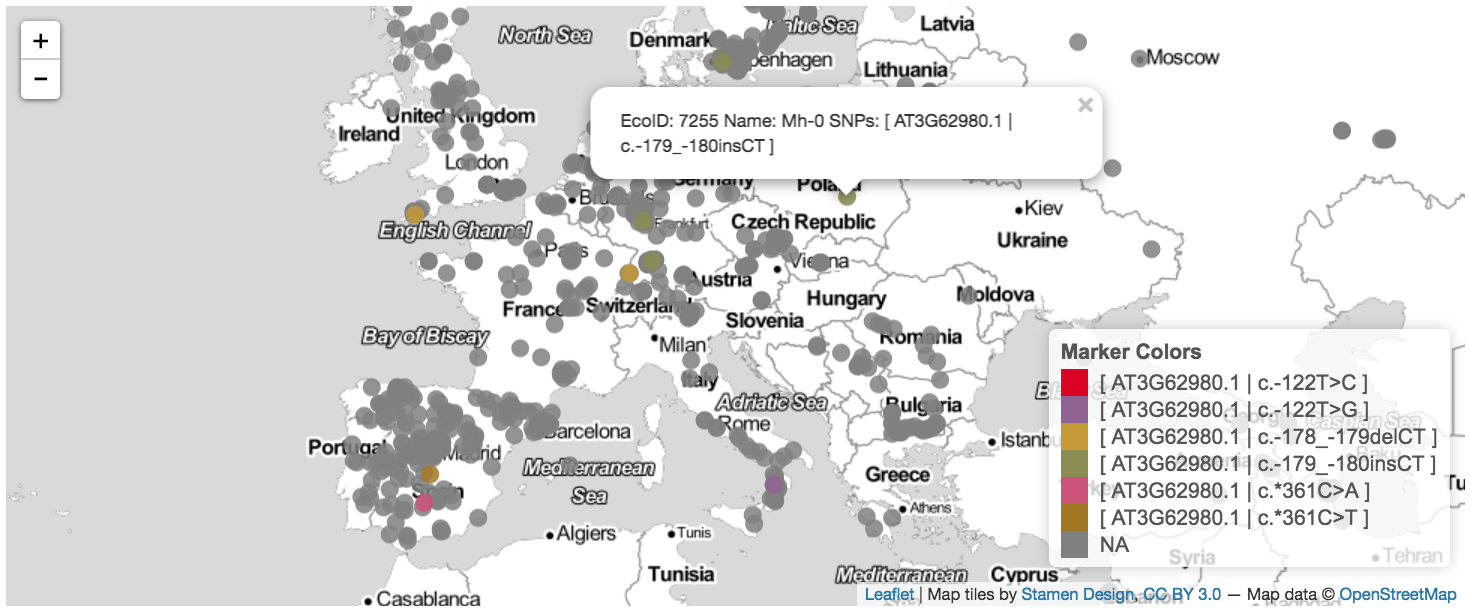
- identify variants/accessions by interacting with these plots, and
- download data for selected sets of variants/accessions.
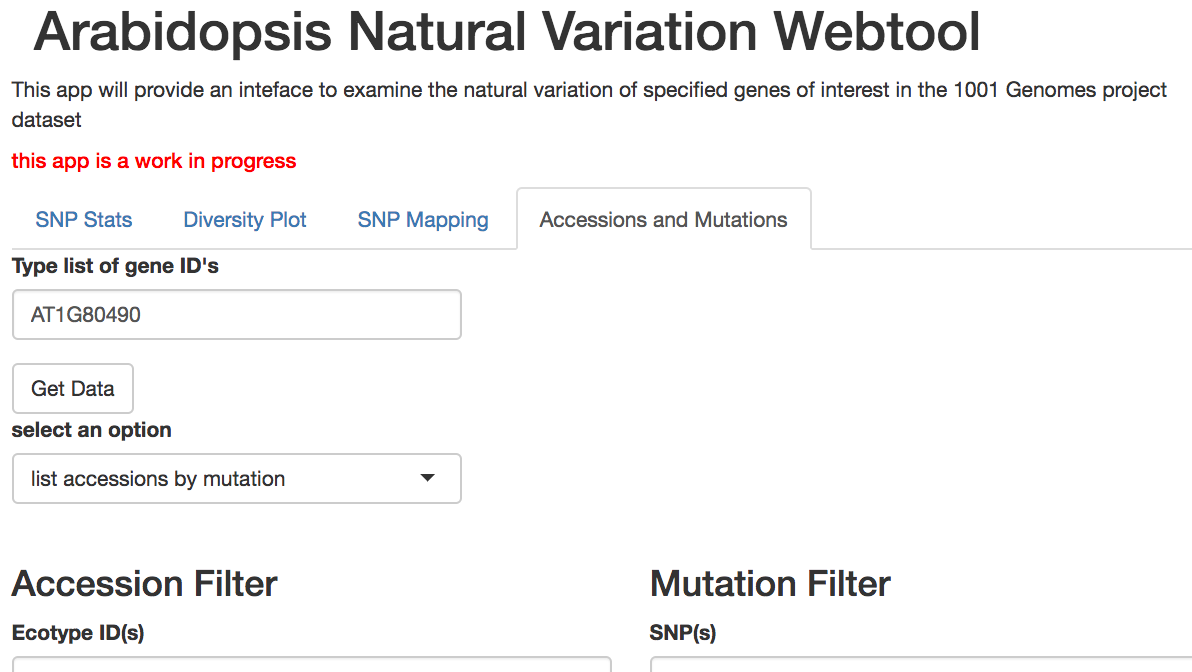
We are currently testing this tool by creating summaries of the natural variation in the genes of nuclear auxin response pathway, and plan to publish this tool and our analysis in early 2018.